Project name: gigas-WGBS-ploidy-desiccation
Funding source: unknown
Species: Crassostrea gigas
variable: ploidy, desiccation, high temperature
Background:
This is continuation of the WGBS analysis I’ve been running on Ronit’s data. The previous post can be found here. I’ll be following Yaamini’s walkthrough in this post to look at the results of bismark.
List of the progress so far:
- Completed bismark on mox. The output was then transferred to gannet by running the rsync command as outlined in the previous post. The output is available here.
- The MultiQC report can be found [here](https://gannet.fish.washington.edu/panopea/030521-ronrosM/multiqc_report.html.
Goals for today:
- Look at MultiQC report.
- Determine effect of ploidy of %mCpG
Step 1: Look at MultiQC report
Looking over the MultiQC report, alignment was amazingly consistent across samples (~61%).
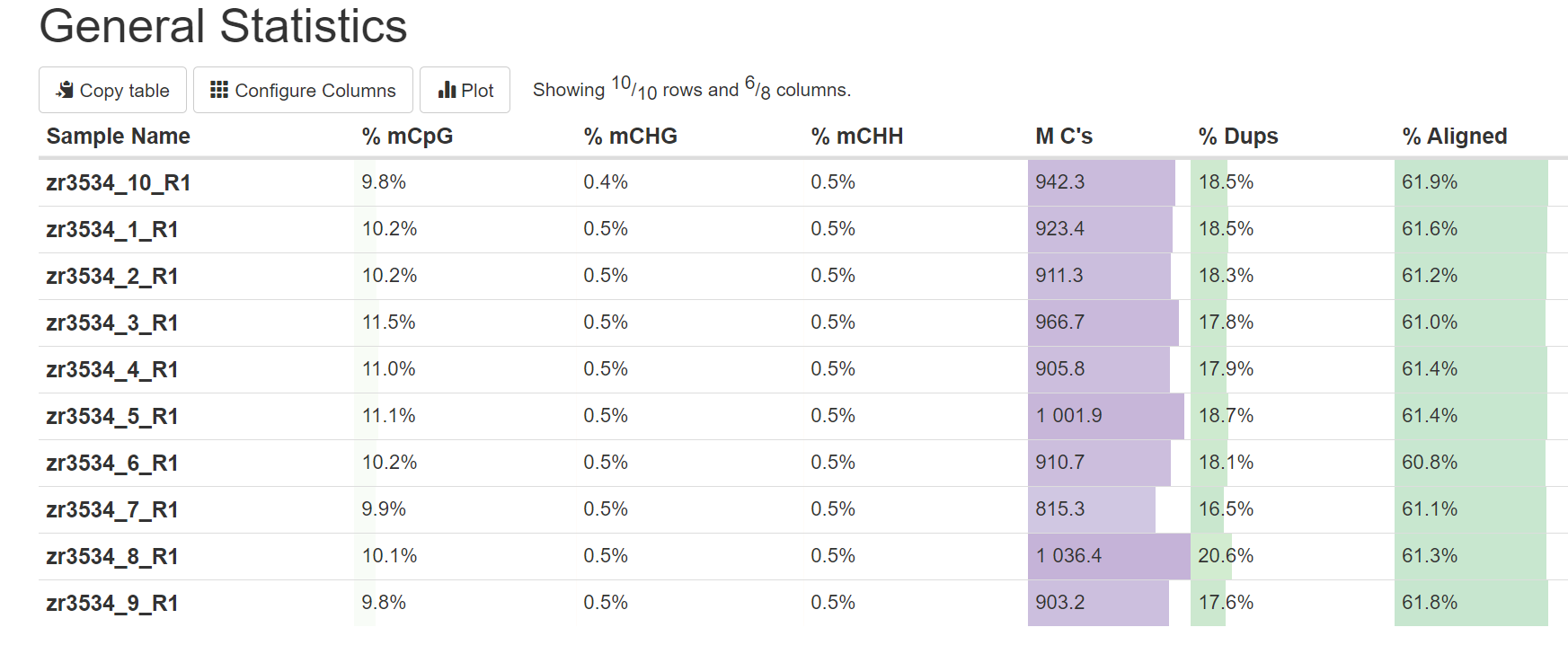
As the samples are from diploid and triploid oysters, a possible issue could differences in genome size across samples. However, the number of reads across all the samples look fairly consistent (68-80 million), with the exception of maybe sample 8 which had almost 90 million.
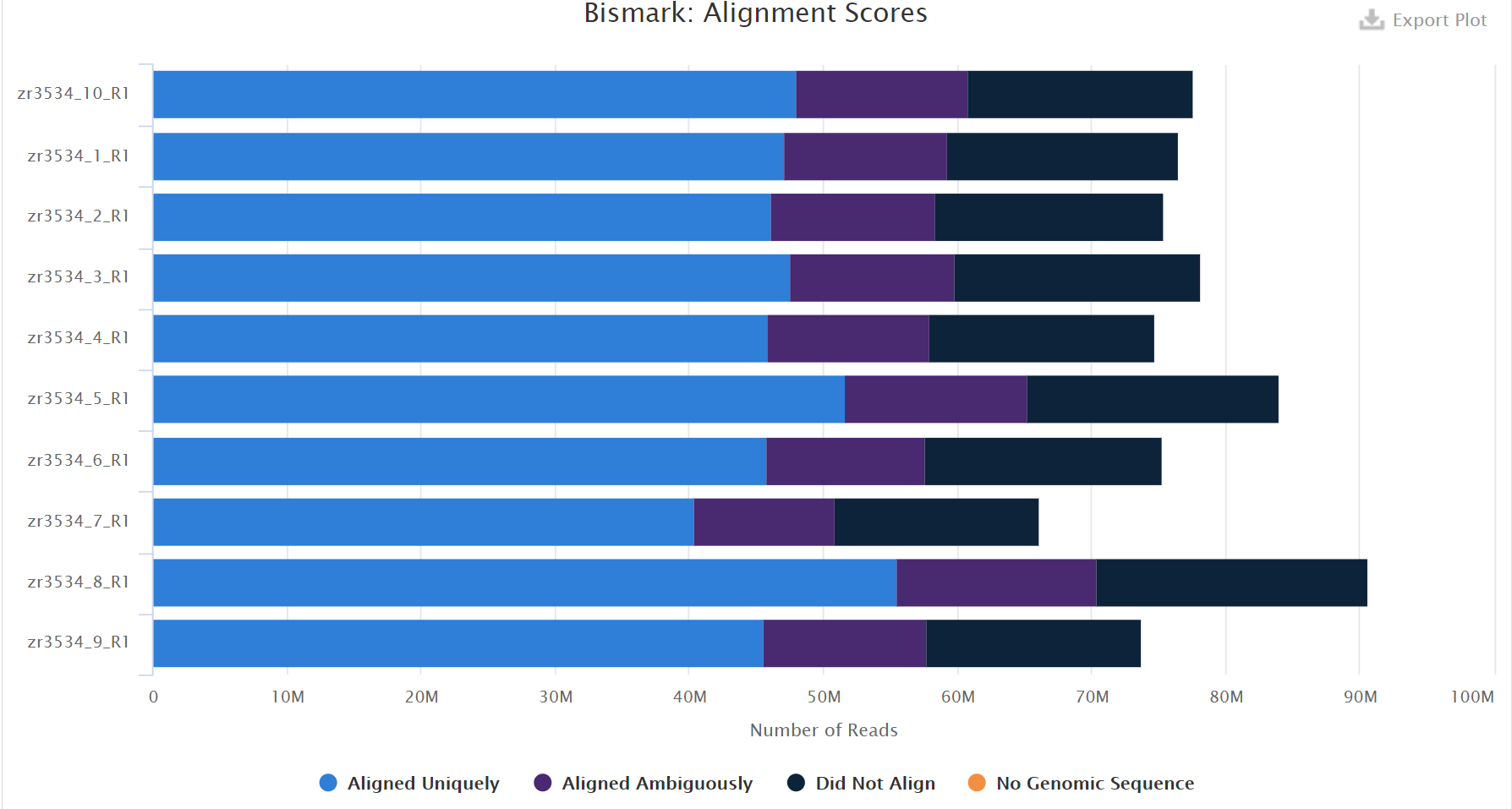
The deduplication percentages were pretty consistent across samples, with the highest being in sample 8 at 20.6%. At first glance there doesn’t seem to be a treatment level effect.
Step 2: Determine effect of ploidy of %mCpG
Previously, Ronit and Shelly used the MethylFlashGlobalDNA Elisa kit to analyze 5-methylcytosine (5-mC) levels in the diploid and triploid cgigas after desiccation stress. Their data and analyses can be found here. After playing around with their R script, it looks like they found significant differences between % 5-mC levels across ploidy (p=0.033), desiccation (p=0.001), as well as an significant ploidy:desiccation interaction (p=0.036).
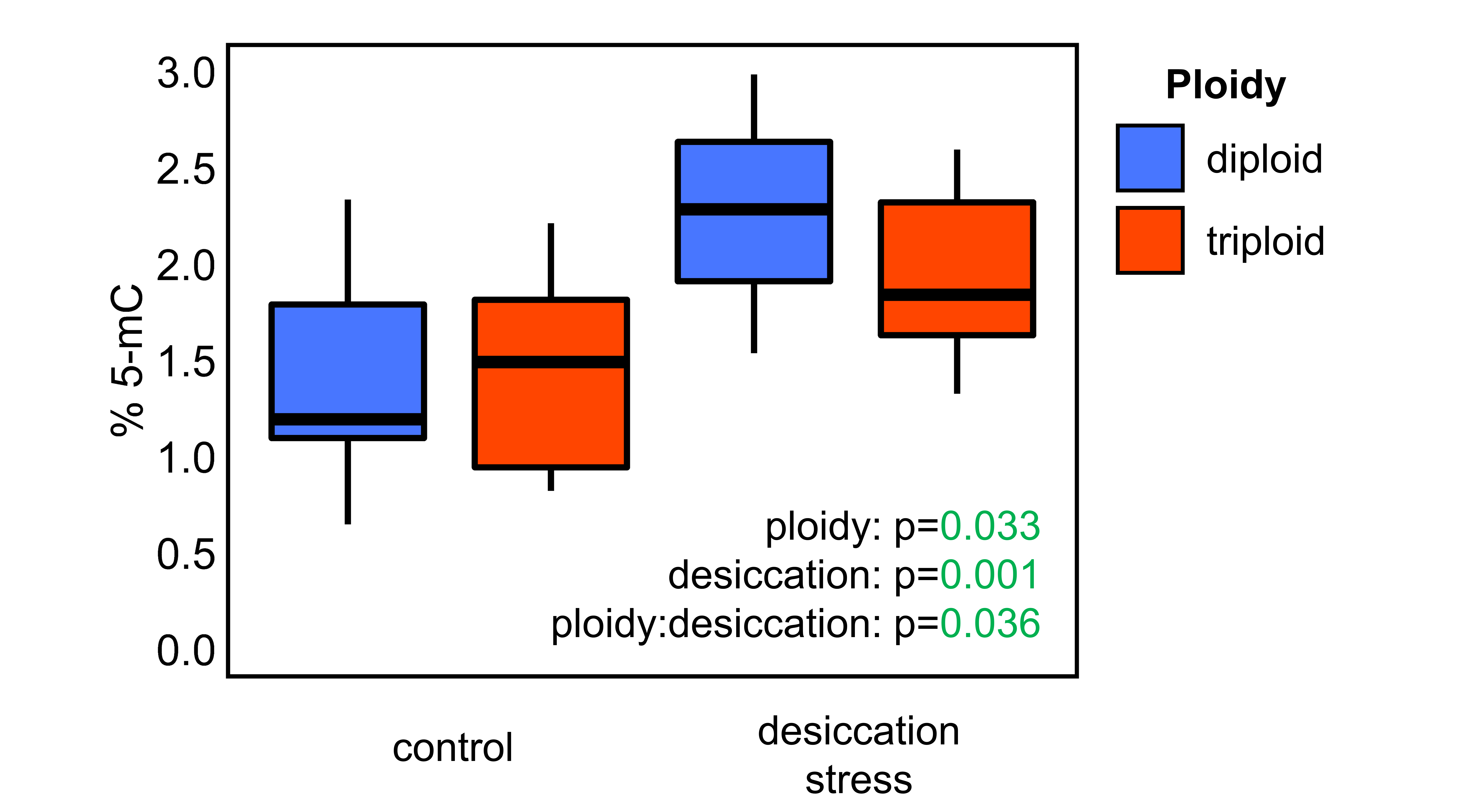
Following up on this result, I analyzed the results of bismark run on 5 oysters from each ploidy exposed to desiccation at 27C for 24 hours. The dataset is further complicated by the fact that two out of each set of 5 animals were further subjected to a 45C shock for 1 hour following desiccation.
The github repo with the WGBS data and analyses can be found here. I ran this R script to analyze the resulting % methylation within CpG islands as reported by MultiQC.
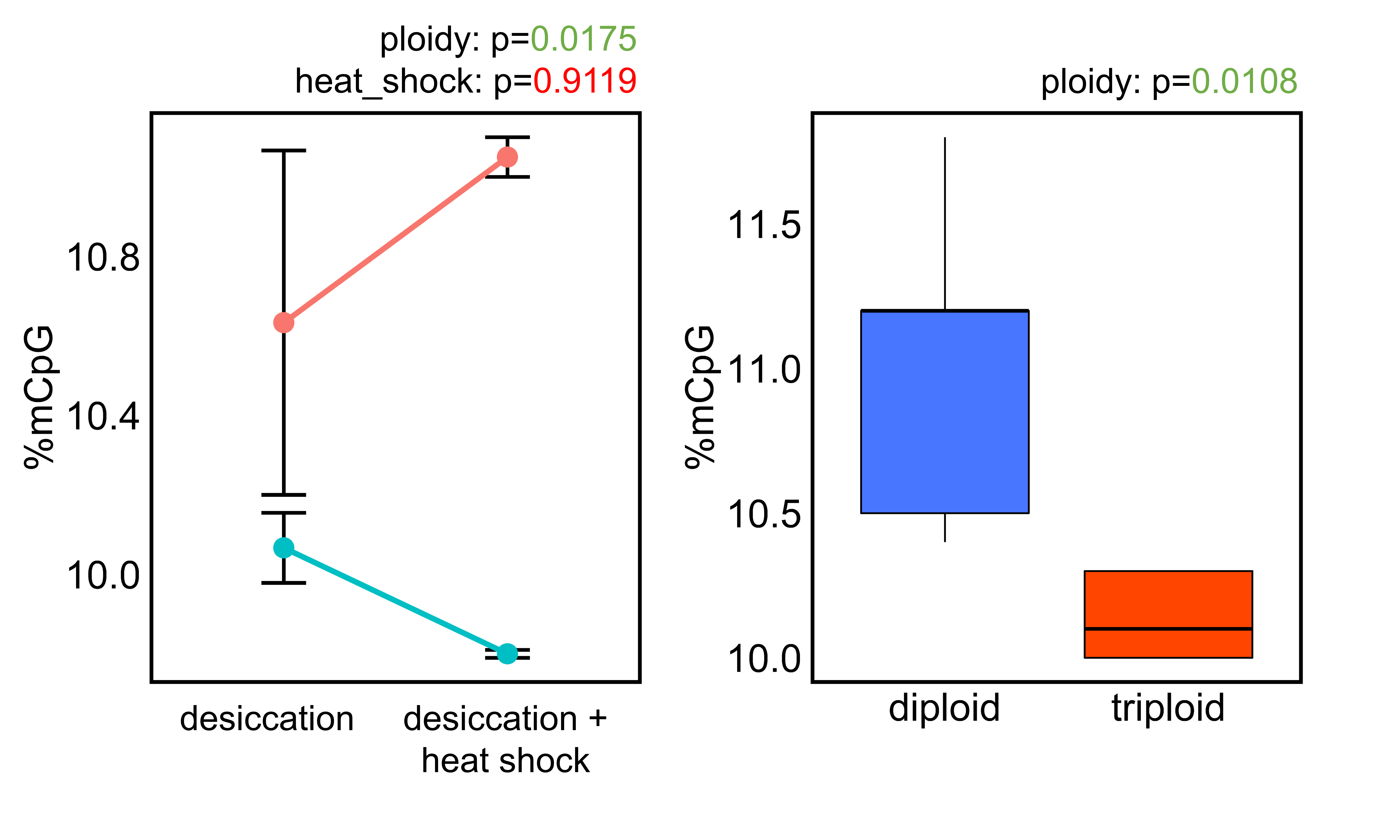
One limitation of this dataset is I don’t have any unstressed animals for comparison. However, it appears ploidy was a significant factor that impacted %mCpG after desiccation (p=0.0175), while their was no measurable effect of subsequent heat shock (p=0.9119). With this result, I lumped together the two groups to generate the second figure, comparing %mCpG expression after desiccation stress (p=0.0108; n=5 per group).
Pathway forward:
- Continue analysis with MethylKit to identify differentially methylated loci (DMLs).