Project name: PSMFC-mytilus-byssus-pilot
Funding source: Pacific States Marine Fisheries Commission
Species: mytilus galloprovincialis, mytilus trossulus
variable: OA, DO, seawater temperature, desiccation
« previous notebook entry « | » next notebook entry »
RNA-seq for de novo transcriptome assembly
We are submitting pooled samples from foot and gill tissue for whole transcriptome sequencing
The mytilus trossulus genome has a c-value of 1.51 according to genomesizes.com
Convert to bp:
1.51 * 0.978 * 10^9 = 1,476,780,000 bp or 1,476 Mbp
Given that approximately 5% of the genome is expressed at any given times in the human genome, I estimate that the transcriptome size is approximately:
1,476,780,000 * 0.05 = 73,839,000 or ~ 74 Mbp
Samples will be submitted to the UT Austin GSAF and run on individual lanes on the NovaSeq S1 PE150, which generates 7*10^8 reads per lane. This yield the following coverage:
700,000,000 bp / 74,000,000 bp = ~10x coverage
Samples for submission. 71 foot samples (from 59 mussels) and 60 gill samples (from 60 mussels) were pooled. The resulting RNA concentrations were 65 ng/ul (Foot - MTF) and 73 ng/ul (Gill - MTG) using the Qubit.
Here are the nanodrop results. Both have a 260/280 ratio of ~2 and the tails are clean, which indicates pure RNA. The Qubit concentrations are more accurate.
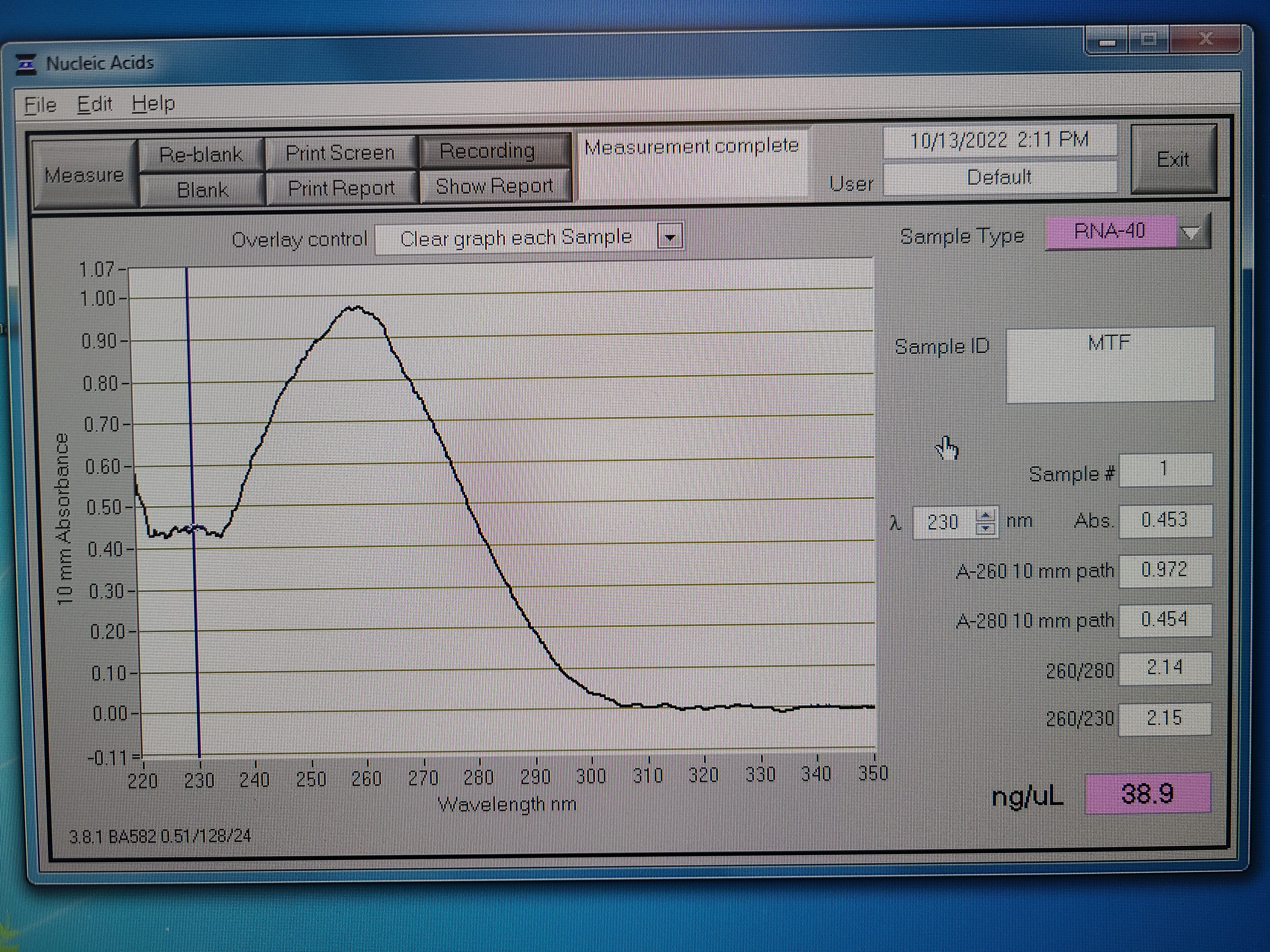
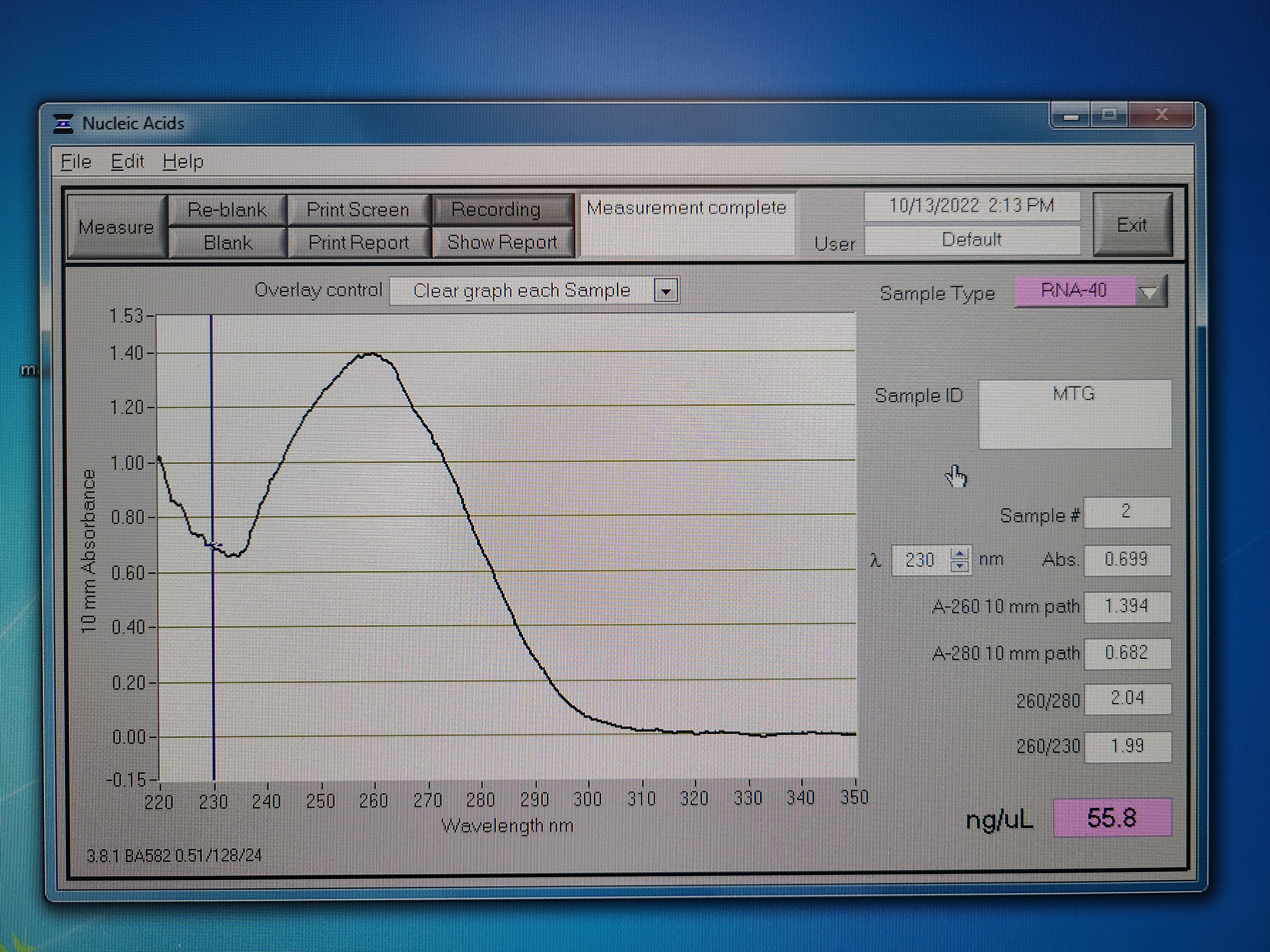
100 ul of each sample were submitted to the GSAF on 10/18/2022.